Education:
B.S. Chemistry, University of Virginia, 2006
Ph.D. Chemistry, Johns Hopkins University, 2015
Professional Experience:
Assistant Professor, University of Missouri, 2021-
Assistant Scientist, University of Wisconsin, 2020-2021
Postdoctoral Fellow, University of Wisconsin, 2015-2020
Honors and Awards:
Ruth L. Kirschstein National Research Service Award (NIH Postdoctoral Fellowship), 2017-2018
Research in the Outlaw Lab lies at the intersection of organic synthesis, chemical biology, and biophysics. Using a combination of techniques from these fields, we seek to create molecular architectures as tools to study and therapeutics to treat human disease.
Current areas of focus include:
1) Development of synthetic methodology to access new molecular scaffolds
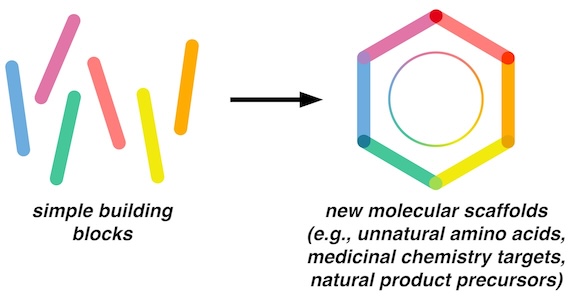
Aromatic and heteroaromatic structures are well-represented in bioactive natural products and medicinal chemistry scaffolds. The substitution pattern tolerance and functional group compatibility afforded by traditional synthetic methods, however, limits the chemical space available to researchers exploring these structures. The Outlaw Lab will develop new synthetic methodology to access aromatic and heteroaromatic scaffolds with substitution patterns and functional group tolerance not available with current methods.
2) Design of synthetic peptides with enhanced properties
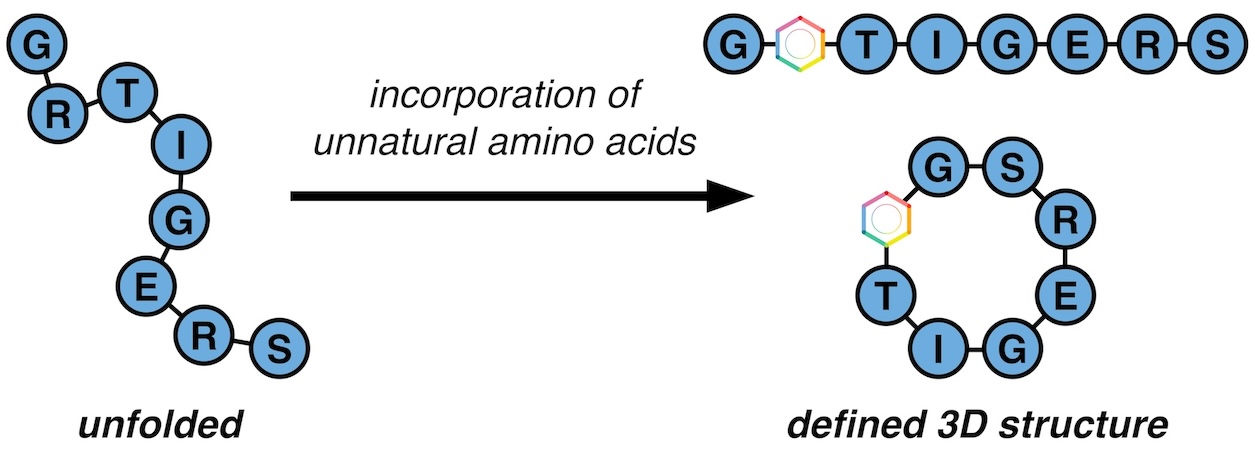
Proteins are made up of polypeptide sequences that adopt three-dimensional structures. Peptides that can mimic specific secondary and tertiary structural elements of proteins have utility as model systems or as inhibitors of protein-protein interactions (PPIs). A major barrier to the development of peptides for therapeutic use has been their poor pharmacokinetic properties (e.g., poor cell permeability, rapid proteolytic degradation). The Outlaw Lab will develop strategies to access peptides with defined three-dimensional architectures and enhanced pharmacokinetic properties.
3) Identification and disruption of molecular interactions underlying viral protein function
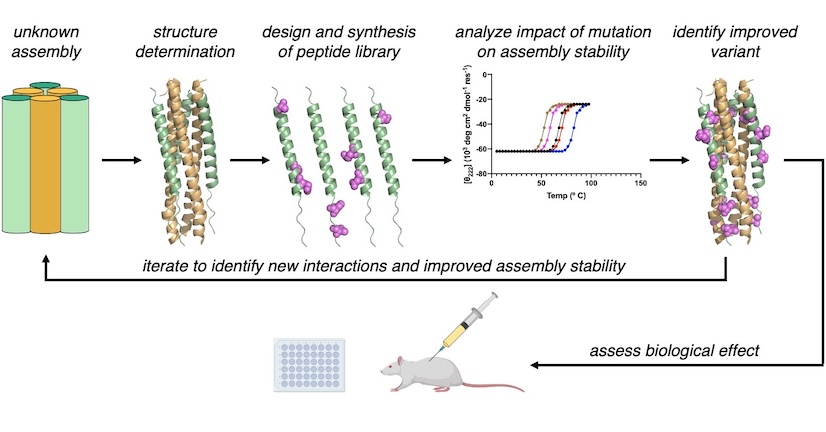
Although the genomes of negative-strand RNA viruses encode only a few proteins, the viruses are able to navigate complex biochemical processes, including fusion with a host cell membrane, replication of the genome, packaging of the genome, evasion of host immune responses, and egress from the host cell. The Outlaw Lab will elucidate the molecular interactions that underly viral protein function and develop new strategies to inhibit these interaction.
- H. Enesi Ozomarisi, K. T. Sharpe, V. K. Outlaw. A Synthetic Route to Highly Substituted 1-Aminonaphthalenes from Readily Available Benzaldehydes. J. Org. Chem. 2024, 89 (2), 1310–1314.
- V. K. Outlaw*, R. W. Cheloha*, E. M. Jurgens*, F. T. Bovier, Y. Zhu, D. F. Kreitler, O. Harder, S. Niewiesk, M. Porotto, S. H. Gellman, A. Moscona. Engineering protease-resistant peptides to inhibit human parainfluenza viral respiratory infection. J. Am. Chem. Soc. 2021, 143 (15), 5958–5966.
*Authors contributed equally - V. K. Outlaw*, F. T. Bovier*, M. C. Mears, M. N. Cajimat, Y. Zhu, M. J. Lin, A. Addetia, N. A. P. Lieberman , V. Peddu, X. Xie, P.-Y. Shi, A. L. Greninger, S. H. Gellman, D. A. Bente, A. Moscona, M. Porotto. Inhibition of coronavirus entry in vitro and ex vivo by a lipid-conjugated peptide derived from the SARS-CoV-2 spike glycoprotein HRC domain. mBio. 2020, 11:e01935-20.
*Authors contributed equally - V. K. Outlaw, S. Bottom-Tanzer, D. F. Kreitler, S. H. Gellman, M. Porotto, A. Moscona. Dual Inhibition of Human Parainfluenza Type 3 and Respiratory Syncytial Virus Infectivity with a Single Agent. J. Am. Chem. Soc. 2019, 141 (32), 12648–12656.
